PROJECTS
AptaSUITE ⇩ BINARY ⇩ SOURCE
AptaSUITE is a platform independent implementation of multiple algorithms designed for the identification of aptamer candidate sequences and the analysis of the SELEX process per se. It provides both, command line and graphical user interfaces.
AptaSUITE is designed to be scalable with both data size and CPU count while minimizing the memory footprint by providing fast, off-heap data structures and storage solutions.
In its core, AptaSUITE consists of a collection of APIs and corresponding implementations facilitating storage, retrieval, and manipulation of aptamer data (such as sequences, aptamer counts in individual selection cycles, structure information and more). On top of these core data structures, a number of previously published algorithms have been implemented. Currently, these are AptaPLEX, AptaSIM, AptaCLUSTER, and AptaTRACE.
Installation and Usage
Download the latest precompiled version from the release page or build the project from source.
git clone https://github.com/drivenbyentropy/aptasuite.git
mvn install
A detailed manual is available through the Wiki pages.
Issues and Comments
If you have any issues or recommendations, please feel free to open a ticket.
Screenshots
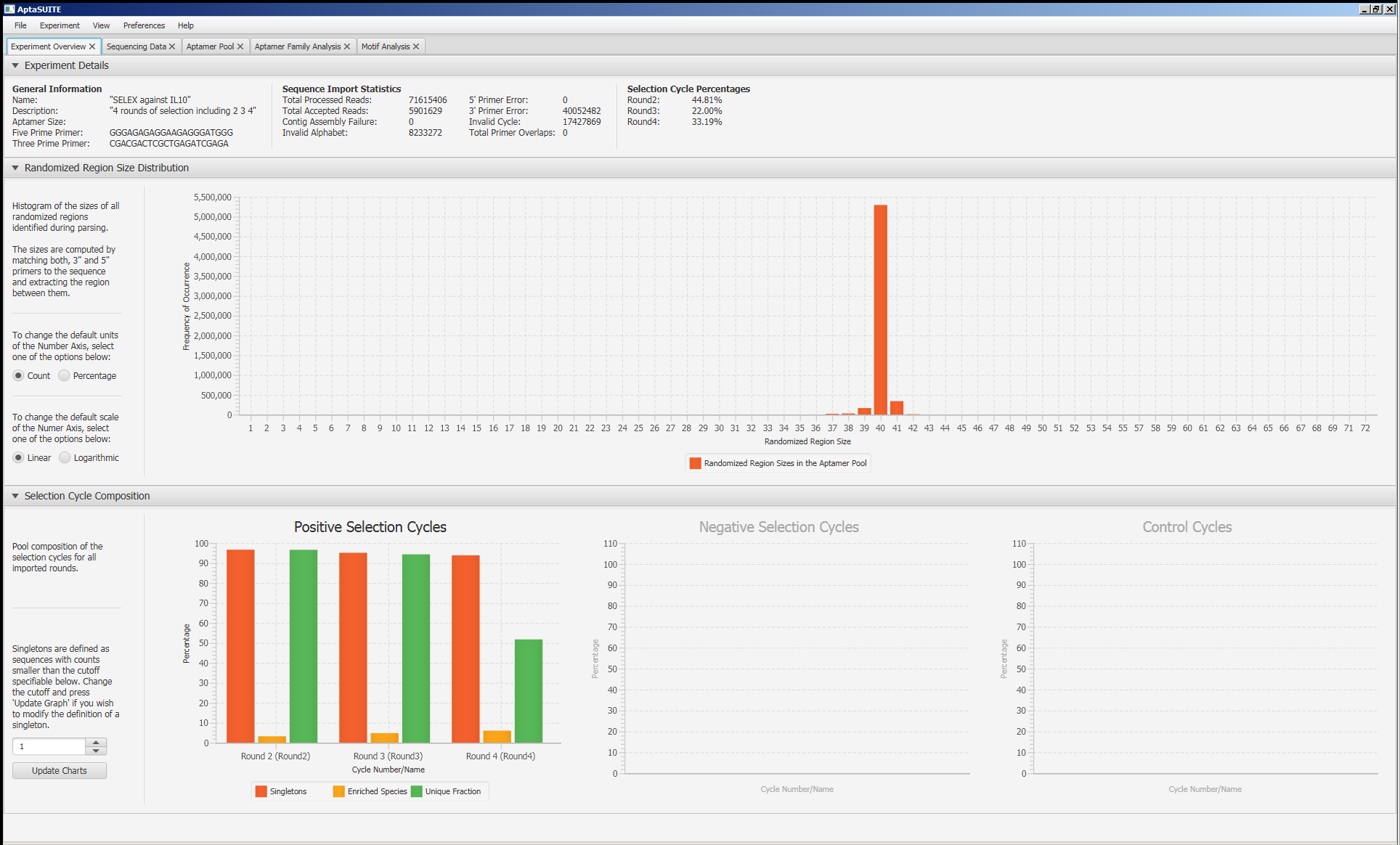
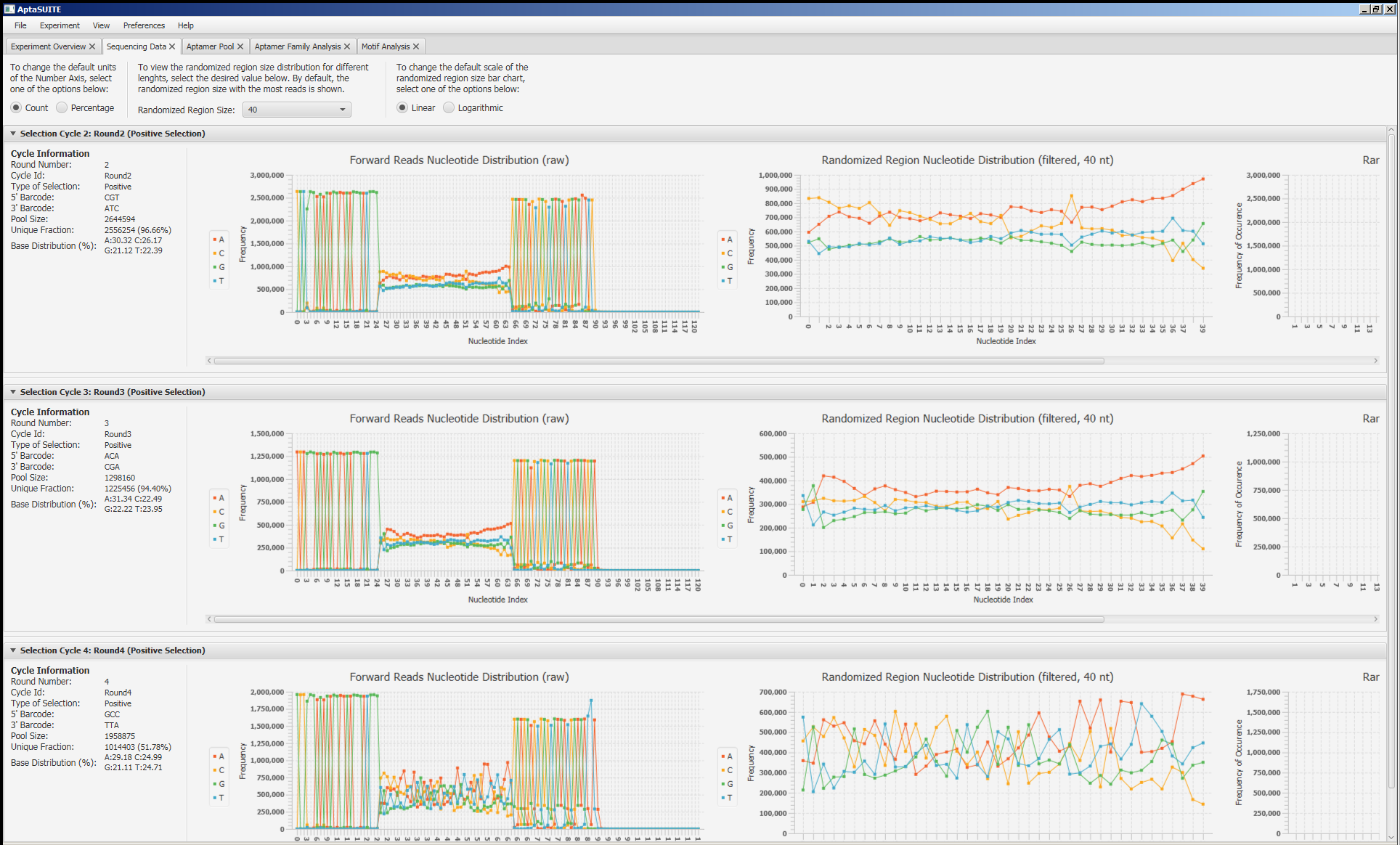
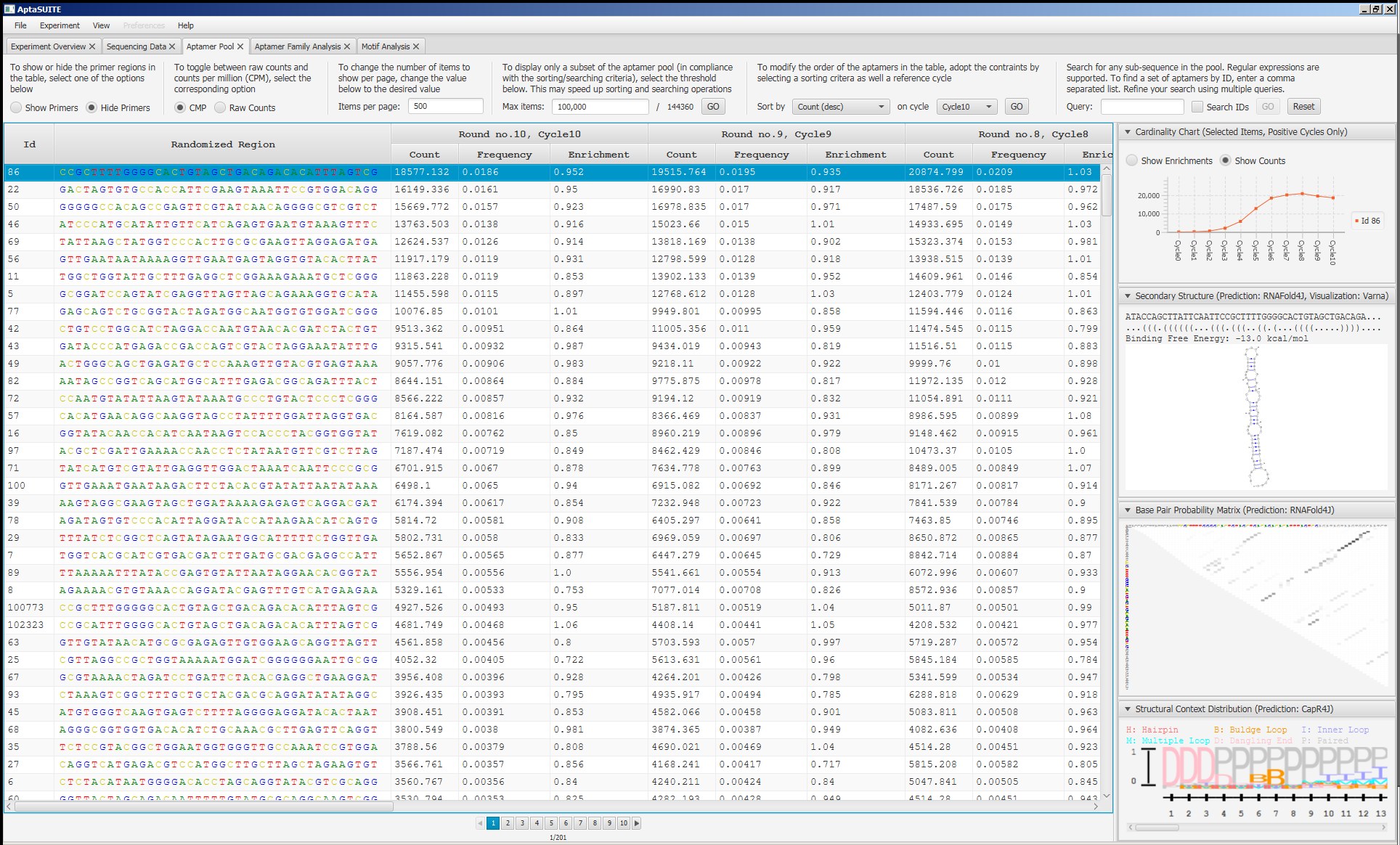

How to Cite
If you use AptaSuite in your work, please cite it as
AptaSUITE: A Full-Featured Bioinformatics Framework for the Comprehensive Analysis of Aptamers from HT-SELEX Experiments.
Hoinka, J., Backofen, R. and Przytycka, T. M. (2018).
Molecular Therapy - Nucleic Acids, 11, 515–517. https://doi.org/10.1016/j.omtn.2018.04.006
I addition, please cite the individual algorithms that aided your analysis. Details on how to cite these can be found in the Help -> How to Cite menu of the graphical user interface.
AptaSUITE is developed and maintained by Jan Hoinka from the AlogCSB lab at the Computational Biology Branch of the National Center for Biotechnology Information / National Library of Medicine, and is released under the GNU Public License (GPL 3.0). We are also grateful for significant ideas and code from Phoung Dao, Rolf Backofen, Teresa Przytycka, and other generous contributors.

